33 DNA fingerprinting, VNTR, Mini satellite and Micro Satellite
1. Introduction
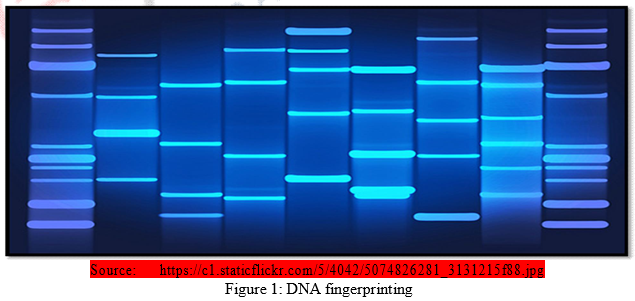
DNA (Deoxyribonucleic acid) fingerprinting is a well known method of identifying criminals. It is considered as recent trend in personal identification. It is a technique to find out variations in individuals of a population at DNA level. The technique of DNA fingerprinting was pioneered and perfected by British geneticist Dr. Alec Jeffreys. In 1984, he had developed this process to visually identify DNA found between the genes in the hope that it will reveal markers for inherited diseases and lead to early treatment. He had no idea at that time that his new technique would help to solve cases related to identity by comparing DNA.
Human “DNA prints” or “DNA fingerprints” are known to possess greater specificity. Like every human organism is unique in terms of their fingerprints, likewise each individual has a unique DNA fingerprint. Unlike conventional fingerprint that occurs only on the fingertips and can be altered by surgery, a DNA fingerprint is the same for the every cell, tissue and organ of a person. It cannot be altered by any known treatment. It represents the blueprint of the individual’s entire genomic makeup.
The “DNA fingerprints”: the specific set of DNA fragments produced by cutting the DNA with a specific set of sequence-specific enzymes, depends on the exact sequence of subunits called nucleotide-pairs in the DNA of the individual. Nucleotides are the molecules that make up DNA. Each nucleotide has a nitrogenous base, a pentose sugar (deoxyribose) and inorganic phosphate. There are double ring purine bases called adenine (A) and guanine (G) and two single ring pyrimidine, cytosine (C) and the thymine (T). This sequence of nucleotide pairs differs in each and every individual. As human genome contains about 3 X 109 nucleotide pairs, it is extremely unlikely that any two individuals other than identical twins will have totally identical “DNA fingerprints.”
DNA fingerprinting can be done by various approaches such as DNA fingerprinting with RFLPs, with PCR, with AmpFLP and with STR. These are the well known methods of DNA finger printing.
1.1 DNA fingerprinting with RFLP (Restriction fragment length polymorphism)
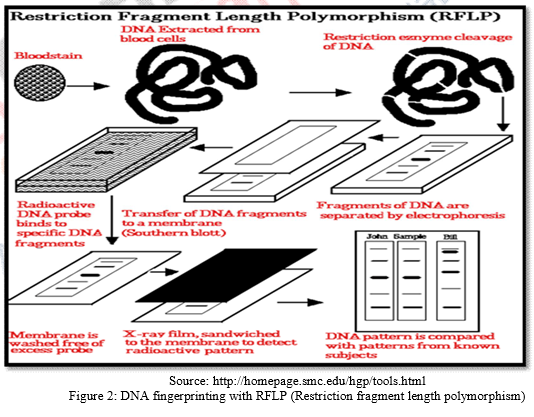
DNA molecules are long double stranded structure tightly packed in chromosomes within the nucleus of each human cell. Within each DNA strand, number of genes are present which determine the specific characteristics of the individual. The genetic information contained in gene composition of DNA fragments are only 5% which are known as coding sequences, and the rest 95% do not contain any genetic information but possess identifiable repetitive sequences of base pairs referred as non- coding sequences. RFLP analyzes the length of the stands of DNA molecules with repetitive nucleotide bases by determining a specific pattern of repeat and then cut those into fragments with specific restriction enzymes. These fragments are then separated by electrophoresis which sorts fragments with respect to length. The segments are than radioactively tagged to produce visual pattern or “DNA fingerprint” on X-ray film. The drawback of this method is that it requires considerable amount of DNA to sort fragments by length.
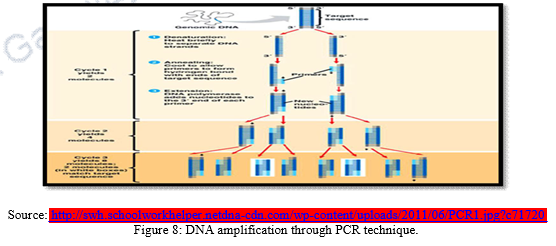
1.2 DNA fingerprinting with PCR (Polymerase chain reaction)
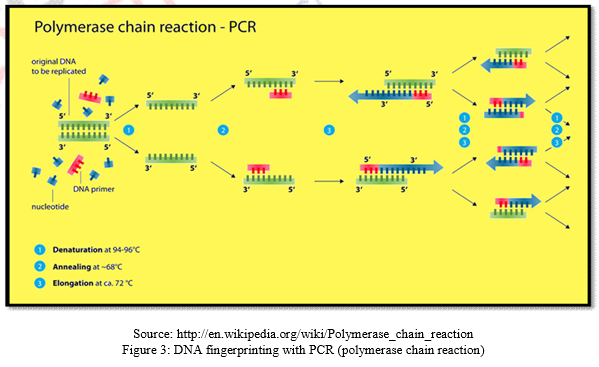
DNA fingerprinting with PCR is most widely used method worldwide. PCR technique was formulated in 1985 by Karry Mullis. The method is designed to permit selective amplification of a specific target DNA sequence within a heterogeneous mixture of DNA sequences. The purpose of a PCR is to make a huge number of copies of a gene. As a result, it now becomes possible to analyze and characterize DNA fragments found in minute quantities in places like a drop of blood at a crime scene or a cell from an extinct dinosaur. Starting with one original copy an almost infinite number of copies can be made using PCR. “Amplified” fragments of DNA can be sequenced, cloned, probed or sized using electrophoresis. Using PCR, defective genes can be amplified to diagnose any number of illnesses. Genes from pathogens can be amplified to identify them (i.e., HIV, Vibrio sp., Salmonella sp. etc.). Amplified fragments can act as genetic fingerprints. PCR technique requires some prior DNA sequence information from the target sequence. From this information, two DNA oligonucleotides primers (amplimers) of about 15 – 25 nucleotides long and specific for the target sequence are designed. The Primers are then added to the denatured template DNA which bind specifically to complementary DNA sequences at the target sites. In the presence of a suitably heat – stable DNA polymerases and DNA precursors (the four deoxynucleoside triphosphates, dATP, dCTP, dGTP, dTTP), primer initiate the synthesis of new DNA strands which are complementary to the individual DNA strands of the target DNA segment. The PCR has a drawback that it cannot discriminate DNA fragments on certain basis as RFLP. PCR technique is very helpful to produce multiple copies of limited DNA molecules.
1.3 DNA fingerprinting with AmpFLP (Amplified fragment length polymorphism)
Amplified fragment length polymorphism or AmpFLP is an attractive version of RFLP as it includes relatively less complicated operation and cost – effective procedure. The method uses PCR technique to amplify the minisatellite loci of the human genome, thus become quicker in recovery than the RFLP. However there are issues to this method about the bunching of the VNTR sequences which is due to use of gel in its analysis phase which ultimately results in misidentification of the DNA finger printing.
1.4 DNA fingerprinting with STR (Short tandem repeats)
STR or Short tandem repeats is another method of DNA fingerprinting which analyzes DNA fragments for the number of repetitions at 13 specific DNA sites. It actually triggers specific areas that have short mini satellites DNA or repeated DNA base pair sequences. STR thus can able to compute how many times a particular sequence of base pair repeat themselves on a particular location within the strand of DNA. This advantage makes the DNA comparisons matching to an almost positive range.
Thus, “DNA fingerprinting” now provide a powerful new tool for establishing identity or non-identity in paternity, rape, and assault cases, as well as other identity cases where tissue samples might be available linking a criminal to the crime. The “DNA fingerprinting” approach has become particularly powerful with the recent development of techniques by which small amount of DNA can be extensively amplified in vitro. This allows “DNA prints” to be made even when only very minute amounts of tissue are available.
2. Principle of DNA fingerprinting
The ideal way to distinguish an individual from other people on Earth would be to describe his or her entire genomic DNA sequence. This idea takes lots of effort, cost, time and thus clearly not practical. So, we look for genes that are highly polymorphic, that is DNA sequences that have multiple alleles in the human population, and therefore different in different individuals. This becomes the principle of DNA fingerprinting or in other words DNA fingerprinting works on the principle of polymorphism in DNA sequences. Important to DNA fingerprinting for determining identity are short nucleotide repeats that vary in number from person to person and are inherited. In human genome about 10% of the entire DNA is formed of repetitive sequences. These sequences are formed of few hundred nucleotides in which a specific sequence of nucleotide bases repeated again and again without interruption and result in the formation of tandem arrangement. These highly repetitive sequences can be categorized as Mini Satellites and Micro Satellites.
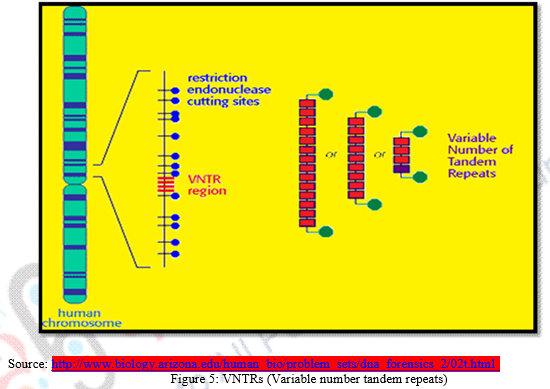
2.1 Mini Satellites DNA & VNTRs
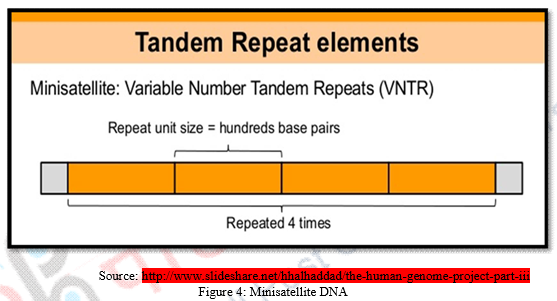
DNA fragments can be categorized as exons or coding sequences that contain genetic information and introns or non-coding sequences which don’t contribute any genetic information about the development of the individual. These introns which seem useless, has been found containing repeated sequences of DNA base pairs. These repetitive sequences are called Mini Satellites. Mini Satellites can be recognized as repetitive DNA fragments that possess 10 – 60 base pair short series of nucleotide bases. In human genome, minisatellites are known to occur at more than 1000 locations. Mini Satellites generally GC rich repetitive sequences, that is sequences with lot of guanine and cytosine base pairs. Length of repeats ranges from 10 to 100 base pair and repeats are tandemly intermingled. When the number of repeats in a given minisatellite varies, we referred it as variable number tandem repeats or VNTRs.
Variable Number Tandem Repeats or VNTRs:
- VNTRs are minisatellite DNA with varied repetitions of specific sequences. In one individual, the specific sequence of mini satellite DNA repeats 5 times whereas in another it may repeat itself 1000 times.
- This makes VNTRs ideal for studying polymorphisms, for linkage analysis as genetic marker, can be implemented as regulators for gene expression, and also used for population studies as VNTRs greatly varies among individuals.
- The VNTRs of two persons may be of the same length and sequence at certain sites, but vary at others sites. This is due to the reason, a child might inherit a chromosome with six tandem repeats from the mother and the same tandem repeated four times in the homologous chromosomes inherited from the father. Thus child’s VNTR alleles resemble half to the mother and half to the father.
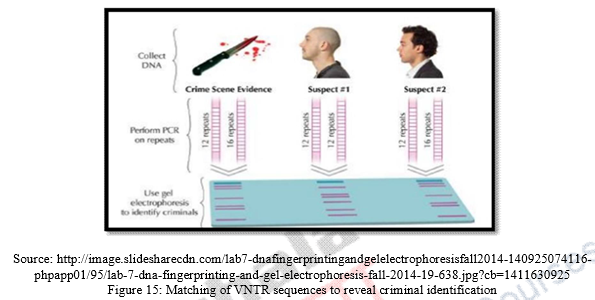
- Every human being has a unique VNTR sequence. A Persons particular VNTR can be determined by performing southern blotting through a hybridization process with radioactive probes that target specific VNTR sequences of DNA fragments will be discussed briefly later in the module.
- VNTR sequences are inherited genetically and an individual possess unique VNTR pattern. While analyzing a individuals VNTR pattern, the use of more and more VNTR probes result in more distinctive and unique individualized pattern of DNA fingerprint which will lead to personal identification of individual in all domain of sciences.
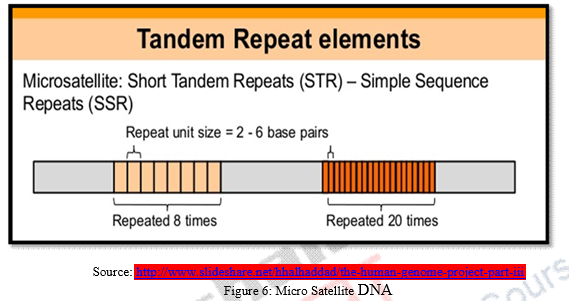
2.2 Micro Satellites
Micro satellites are repetitive DNA fragments that possess 2 – 6 base pairs of nucleotide bases. Micro satellites are generally CA rich repeat tandems with variable number of repeats between alleles which makes it unique as CA nucleotide repeats found very frequently in human genomes and are encountered every thousand base pairs.
Micro satellites are ideal as molecular markers, for determining disputed paternity, for studying gene duplication or deletion, for recombination mapping, for population genetic studies as micro satellites possess locus of many alleles, thus the progenitor of a particular allele or genotypes within pedigree are often be identified. It also provides information regarding closely related alleles. Micro satellites are associated with human diseases. For example, a gene FMR1 has 5 – 60 repeats of a triplet CCG under normal condition in human genome but if the repeat rises above 200, the individual suffered from fragile X syndrome and the individual becomes mentally retarded due to the microsatellite in the fragile site on X chromosome. Some of the other diseases associated with micro satellites are as follows:
- Huntington disease (Gene – HD)
- Fragile X syndrome (Gene – FMR1/FMR2)
- Kennedy disease (Gene – AR)
- Myotonic dystrophy (Gene – DMPK)
- Friedreich ataxia (Gene – X25)
2.3 Significance of Mini & Micro Satellites
Mini satellites and Micro satellites DNA repeat fragments do not have any specific function in DNA structure but their existence is of great importance. Satellite DNAs has crucial role in the construction of genetic maps, isolation of those genes which are responsible for causing human diseases. Technique of DNA finger printing would not have been developed without these satellite DNAs.
3. Technique of DNA fingerprinting
DNA fingerprinting requires only a small amount of tissue like blood or semen or skin cells or even follicle root of the hair. Typically DNA content of about 100,000 cells or about 1 microgram is sufficient.
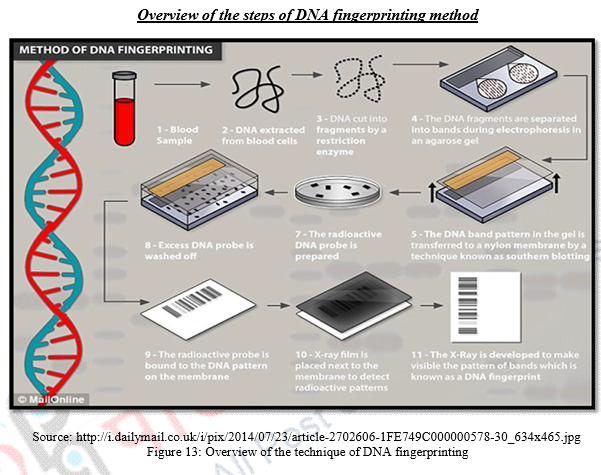
The procedure of DNA fingerprinting involves the following major steps:
a) The DNA is extracted from the nuclei of whatever evidence is availabl For Example, from WBCs in case of blood sample or from hair follicle cells that cling to the roots of hair that have fallen or have been pulled out; or from spermatozoans in semen sample. Nuclei of the cells are extracted in high speed refrigerated centrifuge.
b) If the content of the DNA is limited, DNA can be amplified by making many copies of it using PCR or Polymerase Chain Reaction.
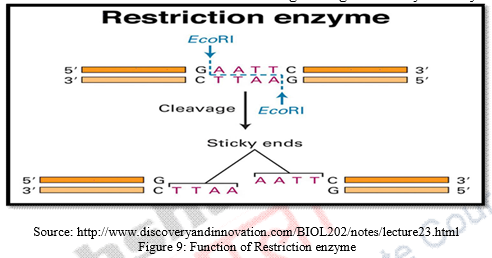
c) The DNA sample so extracted and amplified is digested by a restriction enzyme (EcoRI) which functions to cut the DNA into fragments at specific sites. Thus, these enzymes are site recognizing enzymes for restriction fragment length analysis. The number of these sites present in an individual’s DNA dictates the number and size of DNA fragments generated by the enzymes.
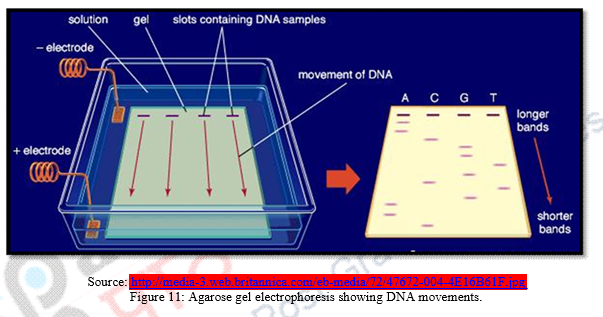
d) Chopped DNA fragments are introduced and passed through electrophoresis set up containing agarose polymer gel. This is done for separation of DNA fragments with respect to their length. The separated fragments can be visualized by staining them with a dye that fluoresces under ultraviolet radiation.
e) In the preceding steps, DNA is in double stranded form. But now double stranded DNA is split into single stranded DNA using alkaline chemicals.
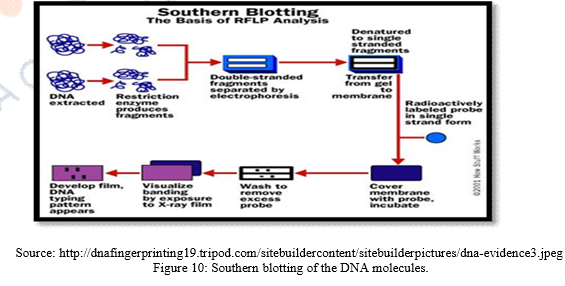
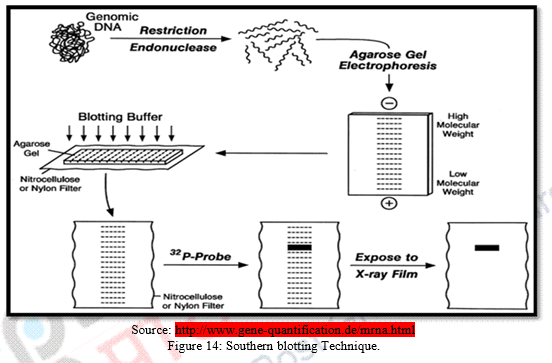
f) These separated DNA sequences are transferred to a nylon or nitrocellulose sheet placed over the gel. This is called “Southern Blotting.”
g) The nylon sheet is then immersed in a bath and probes or markers that are radioactive, synthetic DNA segments of known sequences are The probes target a specific nucleotide sequences which is complementary to the VNTR sequences and hybridize them.
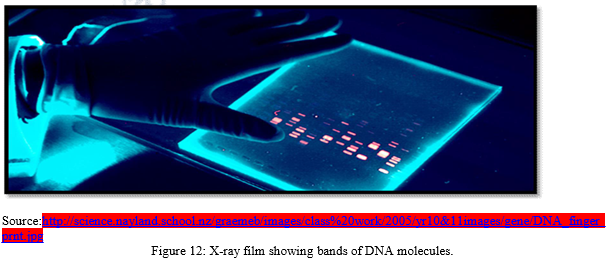
h) Finally X-ray film is exposed to the nylon sheet containing radioactive probes. Dark bands develop at the probe sites which look something like bar codes used to identify items at the grocery store.
Note:
Southern Blotting was named after its inventor Edward M. Southern who developed this method. It is well known technique to detect specific molecules of DNA from a cluster of DNA molecules. It allows investigators to specifically determine the relative molecular weight of a restriction fragment and also it permits to estimate relative amount of DNA molecules in the sample. Scientists use this method to identify sequences of sample DNA molecule by probe hybridization. Procedure of Southern blotting performs by extracting DNA molecule from sample then it is chopped into small fragments with restriction enzymes. Then the cut DNA fragments transferred into agarose gel and split into single stranded DNA molecules by alkaline chemicals. These separated fragments are transferred to a nylon sheet. The sheet is than incubated. Sequences of the single stranded DNA are exposed to known synthetic florescent DNA segments and hybridization occurs and can be viewed through autoradiography on the X-ray film. An adaptation Southern blotting is Northern Blotting where RNA molecules are electrophoresed instead of DNA molecules. Southern blotting has several applications such as identification of desired DNA probes; Detection of mutation; Genetic manipulation can be done. Also it has implication in cloning by determining all the sequences of a sample of DNA or RNA.
4. Application of DNA fingerprinting
- In Forensic Domain:
DNA fingerprinting and forensic science is basically pertains to intersection of law and science. DNA fingerprinting is highly specific and unique. No two individuals have exactly same DNA fingerprints except identical twins. DNA can be isolated from whatever evidence collected at the scene of crime like blood, hair, skin cells, saliva, semen, body tissues. DNA fingerprints thus can be matched or compared with suspected criminals through VNTR prototype.
- Identification of Paternity and maternity:
A child inherits his or her VNTR sequences from his or her parents. Parent- child VNTR sequence analysis can used to solve disputed paternity and maternity cases. The sequences are so unique that parental sequences can be reconstructed even if only the child VNTR sequences are known. This information can also help solve inheritance cases, legal nationality, immigration cases, to verify whether a hopeful immigrant is a really close relative of already an established resident, identity of homicide victim, adoption cases. In 2002, through DNA fingerprinting Elizabeth Hurley prove that Steve Bing was the father of his child Damein.
- Personal identification:
DNA fingerprinting act as a genetic bar code to identify individuals. It is very reliable technique by which personal identification can be made with higher accuracy. It also requires to isolate and analyze VNTR sequences of a particular individual to match it with already registered sequences to conceal identity. Once the identity is assured by fingerprinting technique, identification is 100 percent confirmed or denied.
- Identification of racial groups:
DNA fingerprinting help identify racial groups to rewrite biological evolution.
- Diagnosis of inherited disorders:
DNA fingerprinting is useful in diagnosis of inherited disorders which include Hungtington’s diseases, sickle cell anemia, thalassemia, cystic fibrosis, hemophilia and many more.
- Development of cures for inherited disorders:
DNA fingerprint analysis of relatives who have a history of particular disorder makes it possible to determine sequences of inherited disorders. DNA sequences associated with the disorder can be ascertained and an alternative cure can be developed in the form of DNA sequences with advent technologies and can be implanted to cure the inherited disorder. DNA finger printing was used to identify Chinese medicine – Lingzhi by the Hong Kong Baptist University in 2000.
- Detection of AIDS:
Individuals suffering from HIV – AIDS can be identified by comparing the bands of HIV “RNA” (converted to DNA by RT – PCR) with the bands of the individual DNA isolated from blood sample.
- Breeding program:
DNA fingerprinting permits us to determine the genotype or genome of the individual. Breeders conventionally use DNA fingerprinting to assure homozygous or heterozygous dominance. Offspring of the superior genotype are expected to inherit the highly adaptable characters and DNA fingerprinting allows precise determination of dominant genotype.
5. Summary
DNA fingerprinting is a well known method to determine variation among individuals of population at DNA level. The degree of variation is so high every individual with the exception of identical twins, produce a unique band pattern as every individual has unique set of ordinary fingerprints. It works on the principle of polymorphism in DNA sequences. DNA fingerprinting requires only a small amount of tissue like blood or semen or skin cells or even follicle root of the hair. DNA fingerprinting can be done by various approaches such as DNA fingerprinting with RFLPs, with PCR, with AmpFLP and with STR. These are the well known methods of DNA finger printing. “DNA fingerprinting” is now become a powerful new tool for establishing identity or non-identity in paternity, rape, and assault cases, as well as other identity cases where tissue samples might be available linking a criminal to the crime. The “DNA fingerprinting” approach has become particularly powerful with the recent development of techniques by which small amount of DNA can be extensively amplified in vitro. This allows “DNA prints” to be made even when only very minute amounts of tissue are available.
| you can view video on DNA fingerprinting, VNTR, Mini satellite and Micro Satellite |